Credit Screening Dataset
This dataset has been downloaded from UC Irvine Machine Learning Repository. Link
This dataset is regarding credit card applications.
The target variable/label is whether the application has been granted credit or not.
All attribute names and values have been changed to meaningless symbols to protect
confidentiality of the data.
The objective is here to build a model to give binary output based on the input attributes.
Summary of Key information
Number of Instances/training examples : 690
Number of Instances with missing attributes : 37
Number of qualified Instances/training examples : 653
Number of Input Attributes : 15
Number of categorical attributes : 9
Number of numerical attributes : 6
Target Attribute Type : Binary Class
Target Class distribution : 54%:45%
Problem Identification : Binary Classification with balanced data set
import os
print(os.environ['PATH'])
/usr/local/lib/ruby/gems/3.1.0/bin:/usr/local/opt/ruby/bin:/usr/local/lib/ruby/gems/3.1.0/bin:/usr/local/opt/ruby/bin:/Users/bhaskarroy/opt/anaconda3/bin:/Users/bhaskarroy/opt/anaconda3/condabin:/usr/local/bin:/usr/bin:/bin:/usr/sbin:/sbin:/Library/TeX/texbin:/usr/local/share/dotnet:~/.dotnet/tools:/Library/Frameworks/Mono.framework/Versions/Current/Commands:/usr/local/mysql/bin
from notebook.services.config import ConfigManager
cm = ConfigManager().update('notebook', {'limit_output': 20})
Loading necessary libraries
import numpy as np
import pandas as pd
import time
import seaborn as sns
import matplotlib.pyplot as plt
# Created a custom package named eda and installed it locally
from eda import eda_overview, axes_utils
import category_encoders as ce
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import OrdinalEncoder
from sklearn.model_selection import train_test_split, learning_curve, KFold, cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.svm import LinearSVC
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.metrics import recall_score, precision_score, accuracy_score,confusion_matrix, ConfusionMatrixDisplay, classification_report, f1_score
pd.set_option('display.max_rows', 20)
pd.set_option('precision', 4)
Importing the dataset
path = "/Users/bhaskarroy/BHASKAR FILES/BHASKAR CAREER/Data Science/Practise/" \
"Python/UCI Machine Learning Repository/Credit Screening/"
# Index
# credit.lisp
# credit.names
# crx.data
# crx.names
path1 = path + "crx.data"
path_name = path + "credit.names"
path_crxname = path + "crx.names"
datContent = [i.strip().split() for i in open(path1).readlines()]
len(datContent)
690
print(dir(type(datContent[0][0])))
['__add__', '__class__', '__contains__', '__delattr__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__getnewargs__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__iter__', '__le__', '__len__', '__lt__', '__mod__', '__mul__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__rmod__', '__rmul__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', 'capitalize', 'casefold', 'center', 'count', 'encode', 'endswith', 'expandtabs', 'find', 'format', 'format_map', 'index', 'isalnum', 'isalpha', 'isascii', 'isdecimal', 'isdigit', 'isidentifier', 'islower', 'isnumeric', 'isprintable', 'isspace', 'istitle', 'isupper', 'join', 'ljust', 'lower', 'lstrip', 'maketrans', 'partition', 'replace', 'rfind', 'rindex', 'rjust', 'rpartition', 'rsplit', 'rstrip', 'split', 'splitlines', 'startswith', 'strip', 'swapcase', 'title', 'translate', 'upper', 'zfill']
# Inspecting the contents
print(datContent[0][0].split(sep = ","))
['b', '30.83', '0', 'u', 'g', 'w', 'v', '1.25', 't', 't', '01', 'f', 'g', '00202', '0', '+']
len(datContent[0])
1
Dataset Information
# Opening the file credit.names for the description of data set
with open(path_name) as f:
print(f.read())
1. Title: Japanese Credit Screening (examples & domain theory)
2. Source information:
-- Creators: Chiharu Sano
-- Donor: Chiharu Sano
csano@bonnie.ICS.UCI.EDU
-- Date: 3/19/92
3. Past usage:
-- None Published
4. Relevant information:
-- Examples represent positive and negative instances of people who were and were not
granted credit.
-- The theory was generated by talking to the individuals at a Japanese company that grants
credit.
5. Number of instances: 125
Attributes Information
# Opening the file crx.names for the description of data set
with open(path_crxname) as f:
print(f.read())
1. Title: Credit Approval
2. Sources:
(confidential)
Submitted by quinlan@cs.su.oz.au
3. Past Usage:
See Quinlan,
* "Simplifying decision trees", Int J Man-Machine Studies 27,
Dec 1987, pp. 221-234.
* "C4.5: Programs for Machine Learning", Morgan Kaufmann, Oct 1992
4. Relevant Information:
This file concerns credit card applications. All attribute names
and values have been changed to meaningless symbols to protect
confidentiality of the data.
This dataset is interesting because there is a good mix of
attributes -- continuous, nominal with small numbers of
values, and nominal with larger numbers of values. There
are also a few missing values.
5. Number of Instances: 690
6. Number of Attributes: 15 + class attribute
7. Attribute Information:
A1: b, a.
A2: continuous.
A3: continuous.
A4: u, y, l, t.
A5: g, p, gg.
A6: c, d, cc, i, j, k, m, r, q, w, x, e, aa, ff.
A7: v, h, bb, j, n, z, dd, ff, o.
A8: continuous.
A9: t, f.
A10: t, f.
A11: continuous.
A12: t, f.
A13: g, p, s.
A14: continuous.
A15: continuous.
A16: +,- (class attribute)
8. Missing Attribute Values:
37 cases (5%) have one or more missing values. The missing
values from particular attributes are:
A1: 12
A2: 12
A4: 6
A5: 6
A6: 9
A7: 9
A14: 13
9. Class Distribution
+: 307 (44.5%)
-: 383 (55.5%)
with open(path+"Index") as f:
print(f.read())
Index of credit-screening
02 Dec 1996 182 Index
19 Sep 1992 32218 crx.data
19 Sep 1992 1486 crx.names
16 Jul 1992 12314 credit.lisp
16 Jul 1992 522 credit.names
#with open(path+"credit.lisp") as f:
# print(f.read())
Data preprocessing
Following actions were undertaken:
- Converting to Dataframe Format
- As attribute names are anonymised, create standard feature name starting with ‘A’ and suffixed with feature number
- Handling Missing values : 37 rows had missing values and are not being considered for model building
- Converting Class symbols of Target variable to binary values
- Processing Continous Attributes : Based on inspection, continuous attributes have been converted to float type.
Converting to Dataframe Format
# Inspecting the data
# We find that all the elements in a row is fused as one element.
# We need to use comma for splitting
datContent[0:5]
[['b,30.83,0,u,g,w,v,1.25,t,t,01,f,g,00202,0,+'],
['a,58.67,4.46,u,g,q,h,3.04,t,t,06,f,g,00043,560,+'],
['a,24.50,0.5,u,g,q,h,1.5,t,f,0,f,g,00280,824,+'],
['b,27.83,1.54,u,g,w,v,3.75,t,t,05,t,g,00100,3,+'],
['b,20.17,5.625,u,g,w,v,1.71,t,f,0,f,s,00120,0,+']]
# Splitting using comma to get individual elements
print(datContent[0][0].split(sep = ","))
['b', '30.83', '0', 'u', 'g', 'w', 'v', '1.25', 't', 't', '01', 'f', 'g', '00202', '0', '+']
# The Number of attributes/features is 16
attrCount = len(datContent[0][0].split(sep = ","))
attrCount
16
# As all features names have been changed/anonymised,
# we will create standard feature name starting with 'A' and suffixed with feature number
colNames = ["A"+str(i+1) for i in range(attrCount)]
print(colNames)
['A1', 'A2', 'A3', 'A4', 'A5', 'A6', 'A7', 'A8', 'A9', 'A10', 'A11', 'A12', 'A13', 'A14', 'A15', 'A16']
# Extracting values/data that will be passed as data to create the Dataframe
rawData = []
for i in datContent:
for j in i:
rawData.append(j.split(sep = ","))
# Creating the Dataframe
df = pd.DataFrame(rawData, columns = colNames)
# Inspecting the Dataframe
df.head()
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0 | u | g | w | v | 1.25 | t | t | 01 | f | g | 00202 | 0 | + |
| 1 | a | 58.67 | 4.46 | u | g | q | h | 3.04 | t | t | 06 | f | g | 00043 | 560 | + |
| 2 | a | 24.50 | 0.5 | u | g | q | h | 1.5 | t | f | 0 | f | g | 00280 | 824 | + |
| 3 | b | 27.83 | 1.54 | u | g | w | v | 3.75 | t | t | 05 | t | g | 00100 | 3 | + |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0 | f | s | 00120 | 0 | + |
# Inspecting the dataframe
# We find that features 'A2','A16' have symbols that would require further preprocessing
df.describe()
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 | 690 |
| unique | 3 | 350 | 215 | 4 | 4 | 15 | 10 | 132 | 2 | 2 | 23 | 2 | 3 | 171 | 240 | 2 |
| top | b | ? | 1.5 | u | g | c | v | 0 | t | f | 0 | f | g | 00000 | 0 | - |
| freq | 468 | 12 | 21 | 519 | 519 | 137 | 399 | 70 | 361 | 395 | 395 | 374 | 625 | 132 | 295 | 383 |
# Checking the datatypes to decide the datatype conversions required feature wise
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 690 entries, 0 to 689
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 A1 690 non-null object
1 A2 690 non-null object
2 A3 690 non-null object
3 A4 690 non-null object
4 A5 690 non-null object
5 A6 690 non-null object
6 A7 690 non-null object
7 A8 690 non-null object
8 A9 690 non-null object
9 A10 690 non-null object
10 A11 690 non-null object
11 A12 690 non-null object
12 A13 690 non-null object
13 A14 690 non-null object
14 A15 690 non-null object
15 A16 690 non-null object
dtypes: object(16)
memory usage: 86.4+ KB
Handling Missing values
#df['A2'].astype("float")
df1 = df[(df == "?").any(axis = 1)]
df1
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 71 | b | 34.83 | 4 | u | g | d | bb | 12.5 | t | f | 0 | t | g | ? | 0 | - |
| 83 | a | ? | 3.5 | u | g | d | v | 3 | t | f | 0 | t | g | 00300 | 0 | - |
| 86 | b | ? | 0.375 | u | g | d | v | 0.875 | t | f | 0 | t | s | 00928 | 0 | - |
| 92 | b | ? | 5 | y | p | aa | v | 8.5 | t | f | 0 | f | g | 00000 | 0 | - |
| 97 | b | ? | 0.5 | u | g | c | bb | 0.835 | t | f | 0 | t | s | 00320 | 0 | - |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 608 | b | ? | 0.04 | y | p | d | v | 4.25 | f | f | 0 | t | g | 00460 | 0 | - |
| 622 | a | 25.58 | 0 | ? | ? | ? | ? | 0 | f | f | 0 | f | p | ? | 0 | + |
| 626 | b | 22.00 | 7.835 | y | p | i | bb | 0.165 | f | f | 0 | t | g | ? | 0 | - |
| 641 | ? | 33.17 | 2.25 | y | p | cc | v | 3.5 | f | f | 0 | t | g | 00200 | 141 | - |
| 673 | ? | 29.50 | 2 | y | p | e | h | 2 | f | f | 0 | f | g | 00256 | 17 | - |
37 rows × 16 columns
# Selecting a subset without any missing values
df2 = df[(df != "?").all(axis = 1)]
df2.shape
(653, 16)
df2.head()
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0 | u | g | w | v | 1.25 | t | t | 01 | f | g | 00202 | 0 | + |
| 1 | a | 58.67 | 4.46 | u | g | q | h | 3.04 | t | t | 06 | f | g | 00043 | 560 | + |
| 2 | a | 24.50 | 0.5 | u | g | q | h | 1.5 | t | f | 0 | f | g | 00280 | 824 | + |
| 3 | b | 27.83 | 1.54 | u | g | w | v | 3.75 | t | t | 05 | t | g | 00100 | 3 | + |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0 | f | s | 00120 | 0 | + |
Converting Class symbols of Target variable to binary values
# Below code may return Setting with Copy warning
# Use df._is_view to check if a dataframe is a view or copy
# df2.loc[:, 'A16'] = df2['A16'].map({"-": 0, "+":1}).values
# Use df.assign instead.
# https://stackoverflow.com/questions/36846060/how-to-replace-an-entire-column-on-pandas-dataframe
df2 = df2.assign(A16 = df2['A16'].map({"-": 0, "+":1}))
df2
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0 | u | g | w | v | 1.25 | t | t | 01 | f | g | 00202 | 0 | 1 |
| 1 | a | 58.67 | 4.46 | u | g | q | h | 3.04 | t | t | 06 | f | g | 00043 | 560 | 1 |
| 2 | a | 24.50 | 0.5 | u | g | q | h | 1.5 | t | f | 0 | f | g | 00280 | 824 | 1 |
| 3 | b | 27.83 | 1.54 | u | g | w | v | 3.75 | t | t | 05 | t | g | 00100 | 3 | 1 |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0 | f | s | 00120 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 685 | b | 21.08 | 10.085 | y | p | e | h | 1.25 | f | f | 0 | f | g | 00260 | 0 | 0 |
| 686 | a | 22.67 | 0.75 | u | g | c | v | 2 | f | t | 02 | t | g | 00200 | 394 | 0 |
| 687 | a | 25.25 | 13.5 | y | p | ff | ff | 2 | f | t | 01 | t | g | 00200 | 1 | 0 |
| 688 | b | 17.92 | 0.205 | u | g | aa | v | 0.04 | f | f | 0 | f | g | 00280 | 750 | 0 |
| 689 | b | 35.00 | 3.375 | u | g | c | h | 8.29 | f | f | 0 | t | g | 00000 | 0 | 0 |
653 rows × 16 columns
df2
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0 | u | g | w | v | 1.25 | t | t | 01 | f | g | 00202 | 0 | 1 |
| 1 | a | 58.67 | 4.46 | u | g | q | h | 3.04 | t | t | 06 | f | g | 00043 | 560 | 1 |
| 2 | a | 24.50 | 0.5 | u | g | q | h | 1.5 | t | f | 0 | f | g | 00280 | 824 | 1 |
| 3 | b | 27.83 | 1.54 | u | g | w | v | 3.75 | t | t | 05 | t | g | 00100 | 3 | 1 |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0 | f | s | 00120 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 685 | b | 21.08 | 10.085 | y | p | e | h | 1.25 | f | f | 0 | f | g | 00260 | 0 | 0 |
| 686 | a | 22.67 | 0.75 | u | g | c | v | 2 | f | t | 02 | t | g | 00200 | 394 | 0 |
| 687 | a | 25.25 | 13.5 | y | p | ff | ff | 2 | f | t | 01 | t | g | 00200 | 1 | 0 |
| 688 | b | 17.92 | 0.205 | u | g | aa | v | 0.04 | f | f | 0 | f | g | 00280 | 750 | 0 |
| 689 | b | 35.00 | 3.375 | u | g | c | h | 8.29 | f | f | 0 | t | g | 00000 | 0 | 0 |
653 rows × 16 columns
from eda import datasets
datasets.credit_screening()
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0.000 | u | g | w | v | 1.25 | t | t | 1.0 | f | g | 202.0 | 0.0 | 1 |
| 1 | a | 58.67 | 4.460 | u | g | q | h | 3.04 | t | t | 6.0 | f | g | 43.0 | 560.0 | 1 |
| 2 | a | 24.50 | 0.500 | u | g | q | h | 1.50 | t | f | 0.0 | f | g | 280.0 | 824.0 | 1 |
| 3 | b | 27.83 | 1.540 | u | g | w | v | 3.75 | t | t | 5.0 | t | g | 100.0 | 3.0 | 1 |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0.0 | f | s | 120.0 | 0.0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 685 | b | 21.08 | 10.085 | y | p | e | h | 1.25 | f | f | 0.0 | f | g | 260.0 | 0.0 | 0 |
| 686 | a | 22.67 | 0.750 | u | g | c | v | 2.00 | f | t | 2.0 | t | g | 200.0 | 394.0 | 0 |
| 687 | a | 25.25 | 13.500 | y | p | ff | ff | 2.00 | f | t | 1.0 | t | g | 200.0 | 1.0 | 0 |
| 688 | b | 17.92 | 0.205 | u | g | aa | v | 0.04 | f | f | 0.0 | f | g | 280.0 | 750.0 | 0 |
| 689 | b | 35.00 | 3.375 | u | g | c | h | 8.29 | f | f | 0.0 | t | g | 0.0 | 0.0 | 0 |
653 rows × 16 columns
Processing Continous Attributes
# Continous Variables are A2, A3, A11, A14, A15
contAttr = ['A2', 'A3','A8', 'A11', 'A14', 'A15']
for i in contAttr:
df2.loc[:,i] = df2[i].astype("float")
df2
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | b | 30.83 | 0.000 | u | g | w | v | 1.25 | t | t | 1.0 | f | g | 202.0 | 0.0 | 1 |
| 1 | a | 58.67 | 4.460 | u | g | q | h | 3.04 | t | t | 6.0 | f | g | 43.0 | 560.0 | 1 |
| 2 | a | 24.50 | 0.500 | u | g | q | h | 1.50 | t | f | 0.0 | f | g | 280.0 | 824.0 | 1 |
| 3 | b | 27.83 | 1.540 | u | g | w | v | 3.75 | t | t | 5.0 | t | g | 100.0 | 3.0 | 1 |
| 4 | b | 20.17 | 5.625 | u | g | w | v | 1.71 | t | f | 0.0 | f | s | 120.0 | 0.0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 685 | b | 21.08 | 10.085 | y | p | e | h | 1.25 | f | f | 0.0 | f | g | 260.0 | 0.0 | 0 |
| 686 | a | 22.67 | 0.750 | u | g | c | v | 2.00 | f | t | 2.0 | t | g | 200.0 | 394.0 | 0 |
| 687 | a | 25.25 | 13.500 | y | p | ff | ff | 2.00 | f | t | 1.0 | t | g | 200.0 | 1.0 | 0 |
| 688 | b | 17.92 | 0.205 | u | g | aa | v | 0.04 | f | f | 0.0 | f | g | 280.0 | 750.0 | 0 |
| 689 | b | 35.00 | 3.375 | u | g | c | h | 8.29 | f | f | 0.0 | t | g | 0.0 | 0.0 | 0 |
653 rows × 16 columns
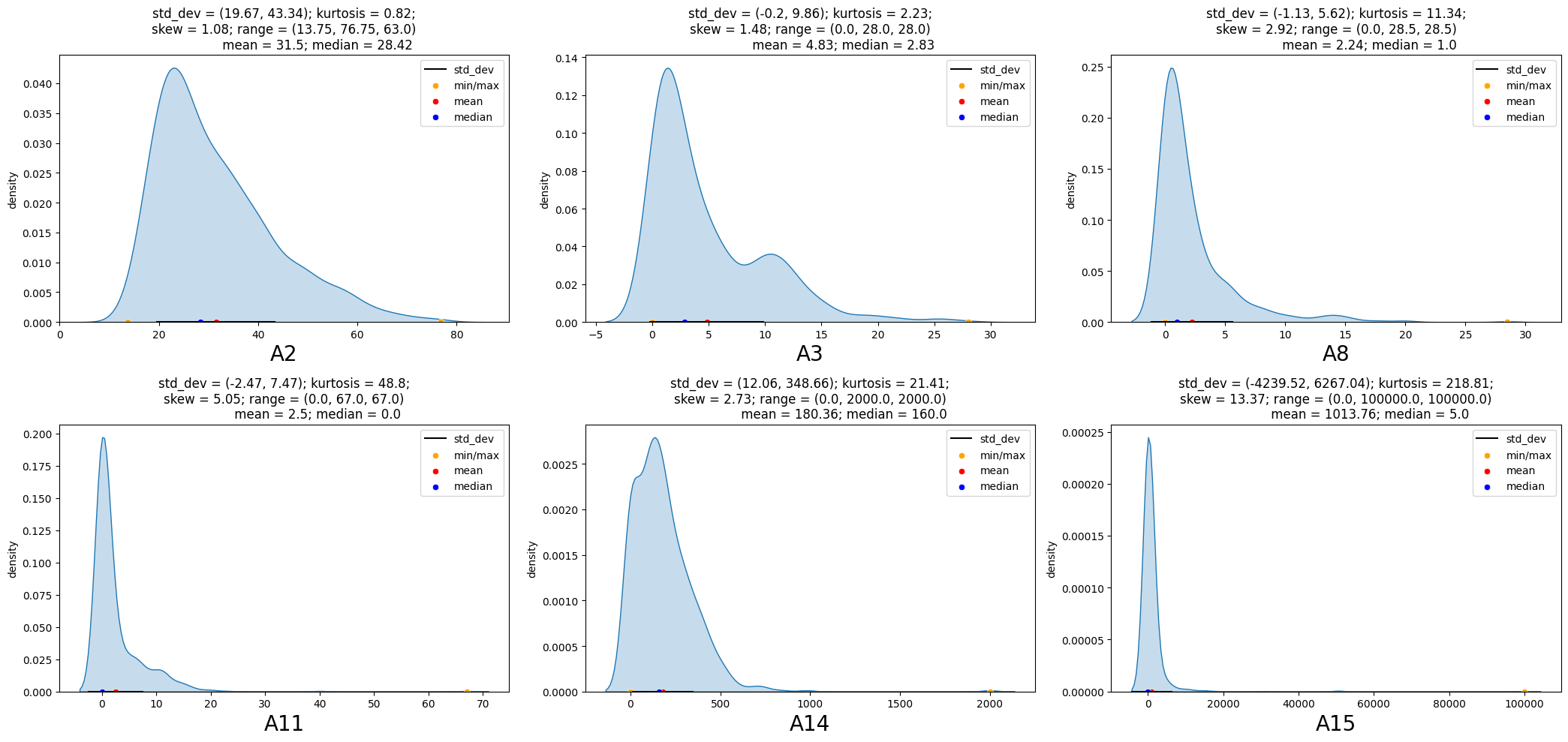
Univariate Analysis - Continous Variables
Findings from the distribution of numeric variables at overall level and considering the application status are as below:
- The dispersion/standard deviation of numeric variables for the applications granted credit extends over a wide range.
- The shape of distribution is similar in both groups for the variables ‘A2’, ‘A3’ and ‘A14’.
- In particular, Numeric variables ‘A11’ and ‘A15’ is concentrated to a very narrow range for the applications not granted credit.
eda_overview.UVA_numeric(data = df2, var_group = contAttr)
# Apply the default theme
sns.set_theme()
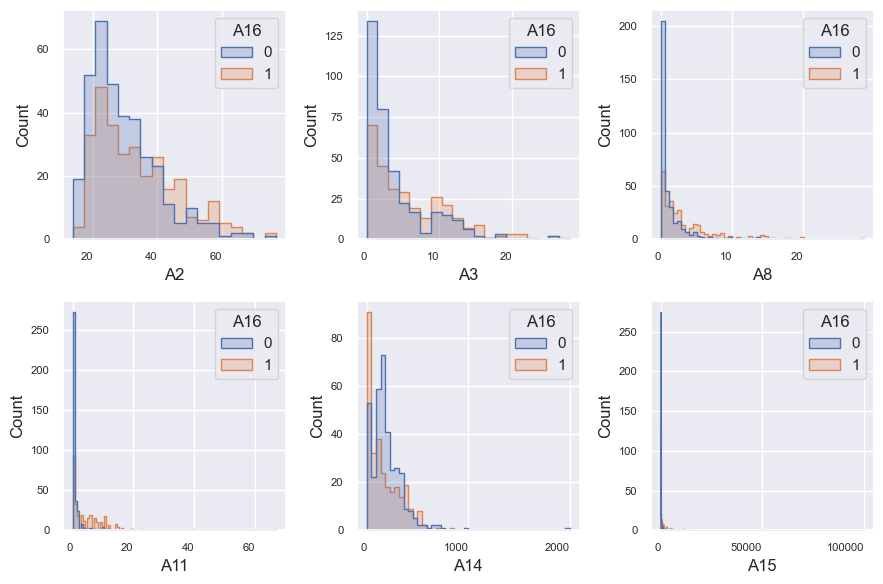
t = eda_overview.UVA_numeric_classwise(df2, 'A16', ['A16'],
colcount = 3, colwidth = 3,
rowheight = 3,
plot_type = 'histogram', element = 'step')
plt.gcf().savefig(path+'Numeric_interaction_class.png', dpi = 150)
t = eda_overview.distribution_comparison(df2, 'A16',['A16'])[0]
t
| Value | Maximum | Minimum | Range | Standard Deviation | Unique Value count | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| A16 category | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Continous Attributes | ||||||||||
| A2 | 74.830 | 76.75 | 15.17 | 13.75 | 59.660 | 63.0 | 10.7192 | 12.6894 | 222 | 219 |
| A3 | 26.335 | 28.00 | 0.00 | 0.00 | 26.335 | 28.0 | 4.3931 | 5.4927 | 146 | 146 |
| A8 | 13.875 | 28.50 | 0.00 | 0.00 | 13.875 | 28.5 | 2.0293 | 4.1674 | 67 | 117 |
| A11 | 20.000 | 67.00 | 0.00 | 0.00 | 20.000 | 67.0 | 1.9584 | 6.3981 | 12 | 23 |
| A14 | 2000.000 | 840.00 | 0.00 | 0.00 | 2000.000 | 840.0 | 172.0580 | 162.5435 | 100 | 108 |
| A15 | 5552.000 | 100000.00 | 0.00 | 0.00 | 5552.000 | 100000.0 | 632.7817 | 7660.9492 | 110 | 145 |
t.to_csv(path +'NumericDistributionComparison.csv')
# Inspecting number of unique values
df2[contAttr].nunique()
A2 340
A3 213
A8 131
A11 23
A14 164
A15 229
dtype: int64
Bivariate Analysis - Continous Variables
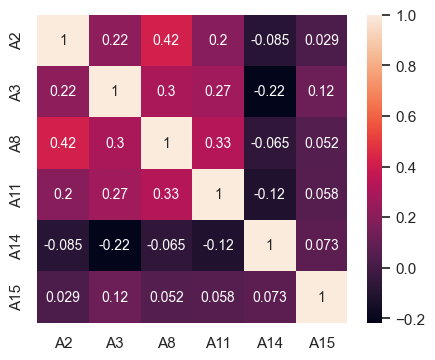
Findings from the correlation plot are as below :
- No significant correlation between any pair of the features
- No significant correlation between any pair of feature and target
# Continous Variables are A2, A3, A11, A14, A15
contAttr = ['A2', 'A3','A8', 'A11', 'A14', 'A15']
# Target Variable is A16
targetAttr = ['A16']
df2[contAttr+targetAttr]
| A2 | A3 | A8 | A11 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|
| 0 | 30.83 | 0.000 | 1.25 | 1.0 | 202.0 | 0.0 | 1 |
| 1 | 58.67 | 4.460 | 3.04 | 6.0 | 43.0 | 560.0 | 1 |
| 2 | 24.50 | 0.500 | 1.50 | 0.0 | 280.0 | 824.0 | 1 |
| 3 | 27.83 | 1.540 | 3.75 | 5.0 | 100.0 | 3.0 | 1 |
| 4 | 20.17 | 5.625 | 1.71 | 0.0 | 120.0 | 0.0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 685 | 21.08 | 10.085 | 1.25 | 0.0 | 260.0 | 0.0 | 0 |
| 686 | 22.67 | 0.750 | 2.00 | 2.0 | 200.0 | 394.0 | 0 |
| 687 | 25.25 | 13.500 | 2.00 | 1.0 | 200.0 | 1.0 | 0 |
| 688 | 17.92 | 0.205 | 0.04 | 0.0 | 280.0 | 750.0 | 0 |
| 689 | 35.00 | 3.375 | 8.29 | 0.0 | 0.0 | 0.0 | 0 |
653 rows × 7 columns
# Bivariate analysis at overall level
plt.rcdefaults()
#sns.set('notebook')
#sns.set_theme(style = 'whitegrid')
sns.set_context(font_scale = 0.6)
from pandas.plotting import scatter_matrix
scatter_matrix(df2[contAttr+targetAttr], figsize = (12,8));
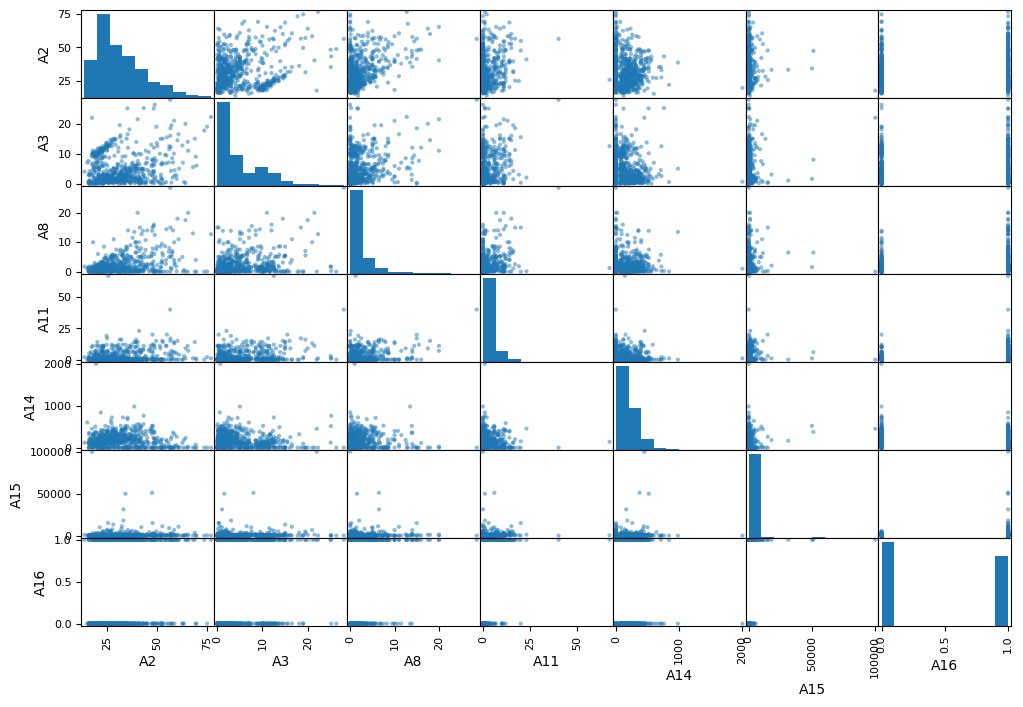
# Bivariate analysis taking into account the target categories
#sns.set('notebook')
sns.set_theme(style="darkgrid")
sns.pairplot(df2[contAttr+targetAttr],hue= 'A16',height = 1.5)
<seaborn.axisgrid.PairGrid at 0x7fc08ae9e790>
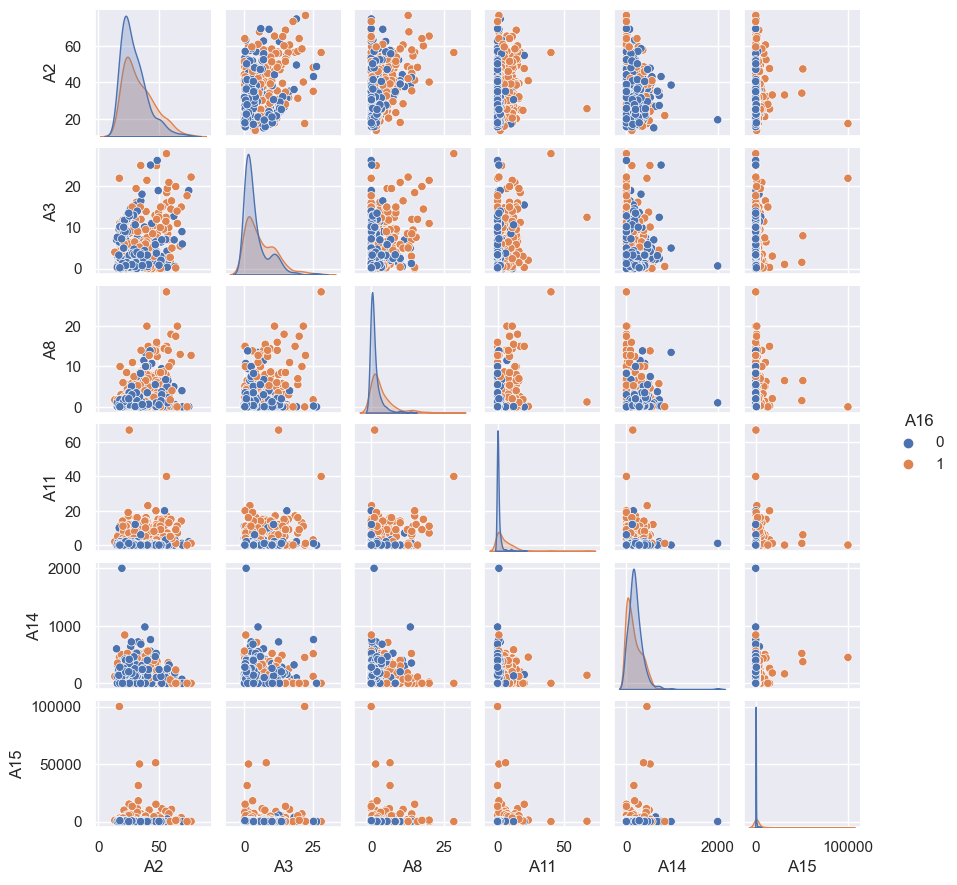
df2[contAttr+targetAttr].dtypes
A2 float64
A3 float64
A8 float64
A11 float64
A14 float64
A15 float64
A16 int64
dtype: object
# Correlation table
df2[contAttr].corr()
| A2 | A3 | A8 | A11 | A14 | A15 | |
|---|---|---|---|---|---|---|
| A2 | 1.0000 | 0.2177 | 0.4176 | 0.1982 | -0.0846 | 0.0291 |
| A3 | 0.2177 | 1.0000 | 0.3006 | 0.2698 | -0.2171 | 0.1198 |
| A8 | 0.4176 | 0.3006 | 1.0000 | 0.3273 | -0.0648 | 0.0522 |
| A11 | 0.1982 | 0.2698 | 0.3273 | 1.0000 | -0.1161 | 0.0584 |
| A14 | -0.0846 | -0.2171 | -0.0648 | -0.1161 | 1.0000 | 0.0734 |
| A15 | 0.0291 | 0.1198 | 0.0522 | 0.0584 | 0.0734 | 1.0000 |
# Heatmap for correlation of numeric attributes
fig, ax = plt.subplots(figsize=(5,4))
sns.heatmap(df2[contAttr].corr(), annot = True, ax = ax, annot_kws={"fontsize":10});
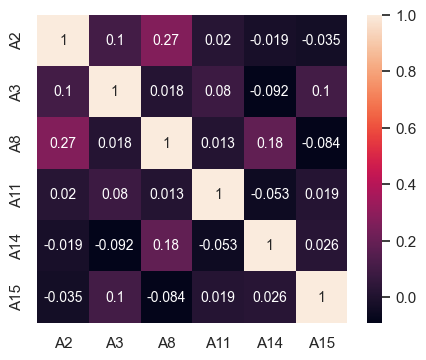
# Correlation matrix for customers not granted credit
fig, ax = plt.subplots(figsize=(5,4))
sns.heatmap(df2[df2['A16'] == 0][contAttr].corr(), ax = ax, annot_kws={"fontsize":10}, annot = True);
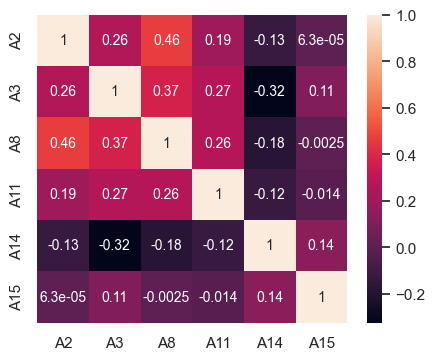
# Correlation matrix for customers granted credit
fig, ax = plt.subplots(figsize=(5,4))
sns.heatmap(df2[df2['A16'] == 1][contAttr].corr(),ax = ax,
annot_kws={"fontsize":10}, annot = True);
Univariate Analysis - Categorical Variables
# Continous Variables are A2, A3, A8, A11, A14, A15
# Categorical Input Variables are A1, A4, A5, A6, A7, A9, A10, A12, A13
# Target Variable is A16 and is categorical.
catAttr = ["A1","A4", "A5", "A6", "A7", "A9", "A10", "A12", "A13"]
eda_overview.UVA_category(df2, var_group = catAttr + targetAttr,
colwidth = 3,
rowheight = 2,
colcount = 2,
spine_linewidth = 0.2,
nspaces = 4, ncountspaces = 3,
axlabel_fntsize = 7,
ax_xticklabel_fntsize = 7,
ax_yticklabel_fntsize = 7,
change_ratio = 0.6,
infofntsize = 7)
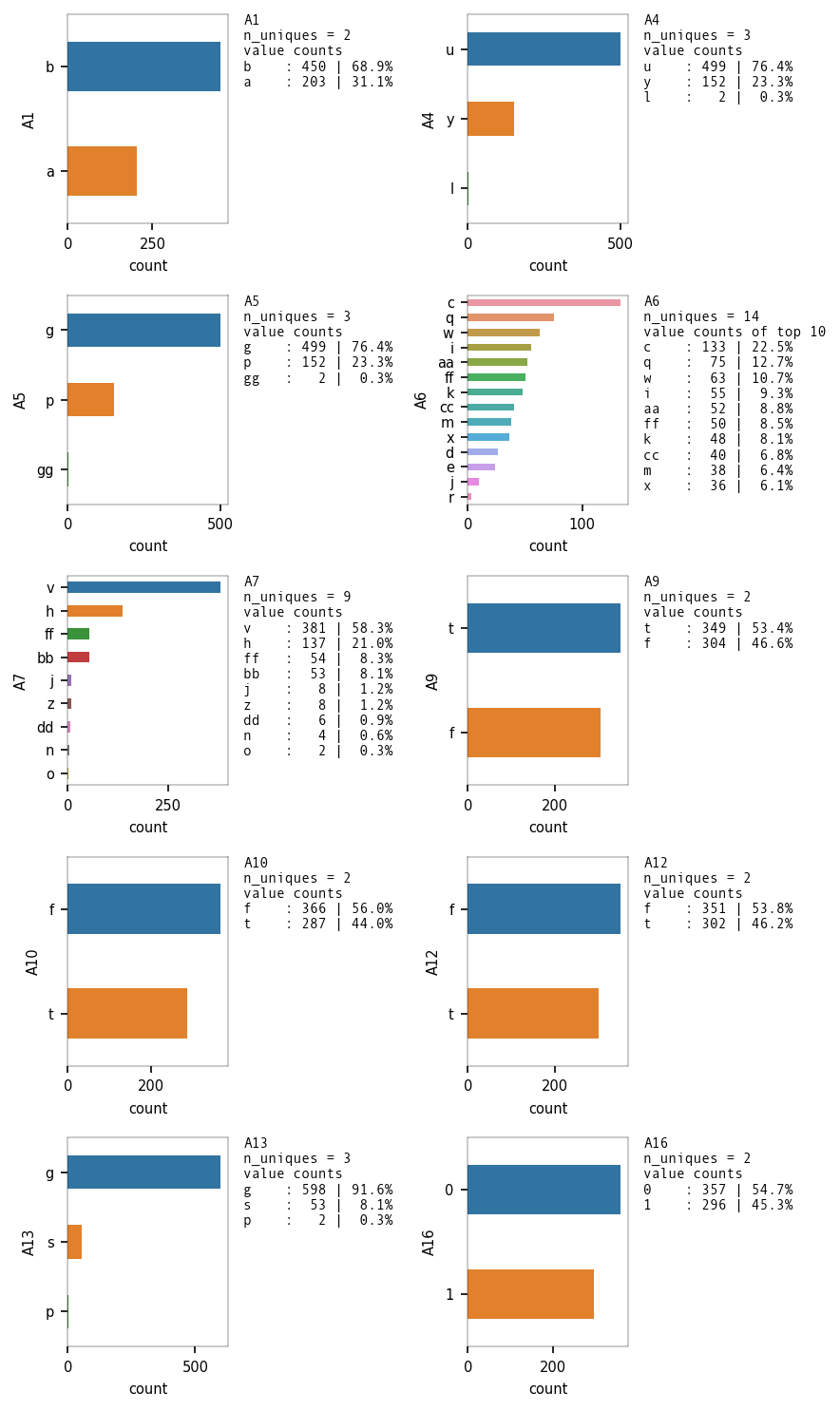
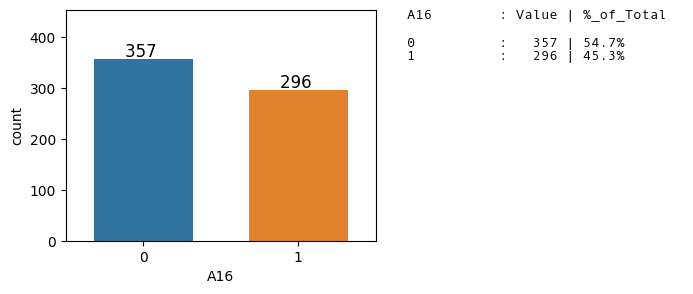
Distribution of the Target Class
Dataset is balanced as the ratio of the binary classes is ~55:45.
We can use Accuracy as a Evaluation metric for the classifier model.
plt.figure(figsize = (4,3), dpi = 100)
ax = sns.countplot(x = 'A16', data = df2, )
ax.set_ylim(0, 1.1*ax.get_ylim()[1])
axes_utils.Add_data_labels(ax.patches)
axes_utils.Change_barWidth(ax.patches, 0.8)
axes_utils.Add_valuecountsinfo(ax, 'A16',df2)